
 |
 |

 |
 |
 |
 |
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
Selected YASARA Screenshots
The screenshots below are taken
directly from the YASARA window, the scenes can be interactively
explored with YASARA's new Vulkan graphics engine, which supports
real-time soft shadows, per-pixel ambient lighting and billions of
atoms. OpenGL is still available for old graphics cards. Many
additional screenshots can be found by following the links below:
- YASARA's best graphics: movies for e-learning tutorials and presentations
- YASARA's look across the various operating systems
- Many more screenshots showing YASARA
features
 Myosin with ADP and some important residues highlighted |
 Interactive visualization of giant structures, each of the 43 million atoms can be clicked, modified or analyzed |
 The genome builder, which takes a FASTA sequence file and nucleic acid binding proteins to build an entire genome, suitable for coarse-grained MD. |
 A molecular dynamics simulation of crambin |
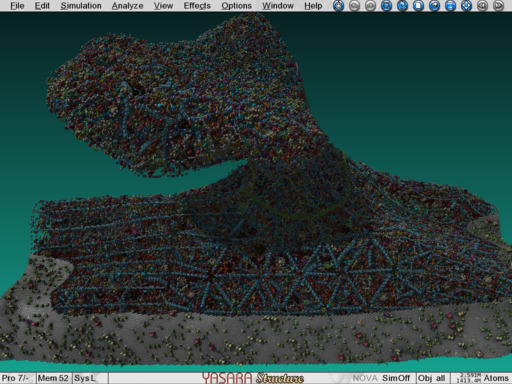 The postsynapse, a gigastructure built with YASARA. You can interactively zoom in and click, modify or analyze each of the 1.4 billion atoms |
 Converting structures to a coarse-grained, rigid representation for faster MD simulation |
 With YASARA you can easily create your custom analysis, write the results to text tables or automatically create scientific reports |
 A sky-sphere in the background, with reflections in protein secondary structure |
 YASARA's mixed multiple timestep algorithm to accelerate molecular dynamics simulations |
 Eight ways to visualize electrostatic potentials from Particle Mesh Ewald (PME) or Poisson-Boltzmann (PBS) calculations |
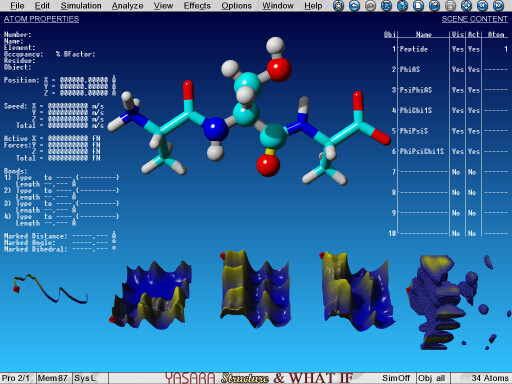 The 1D, 2D and 3D knowledge-based dihedral angle potentials of a serine residue in the YASARA force field |
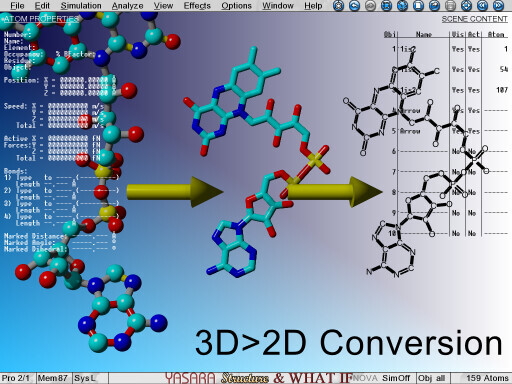 Turning 3D molecules into flat 2D structures for visualization as structural formulas |
 Analyzing cation-Pi interactions |
 The small-molecule builder used to create tiotropium |
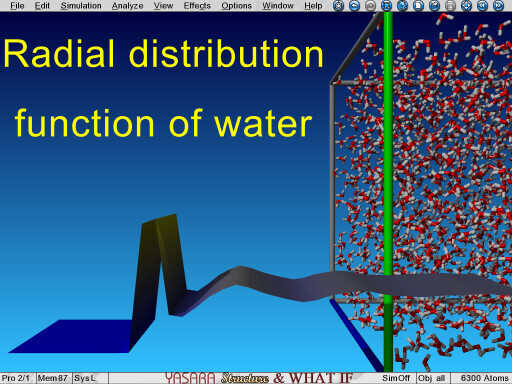 Radial distribution function of water calculated from an MD trajectory |
 Ray-tracing with the built-in POV-Ray in comic style, adding outlines via depth-buffer analysis |
 Ray-tracing with the built-in POV-Ray, adding granite textures |
 A protein crystal including surfaces cut at the periodic boundaries |
 A molecular surface cut open by an interactive clipping-plane |
 An example macro that creates images/textures, shows text and a clickable button on each, and places them in bouncing spheres |